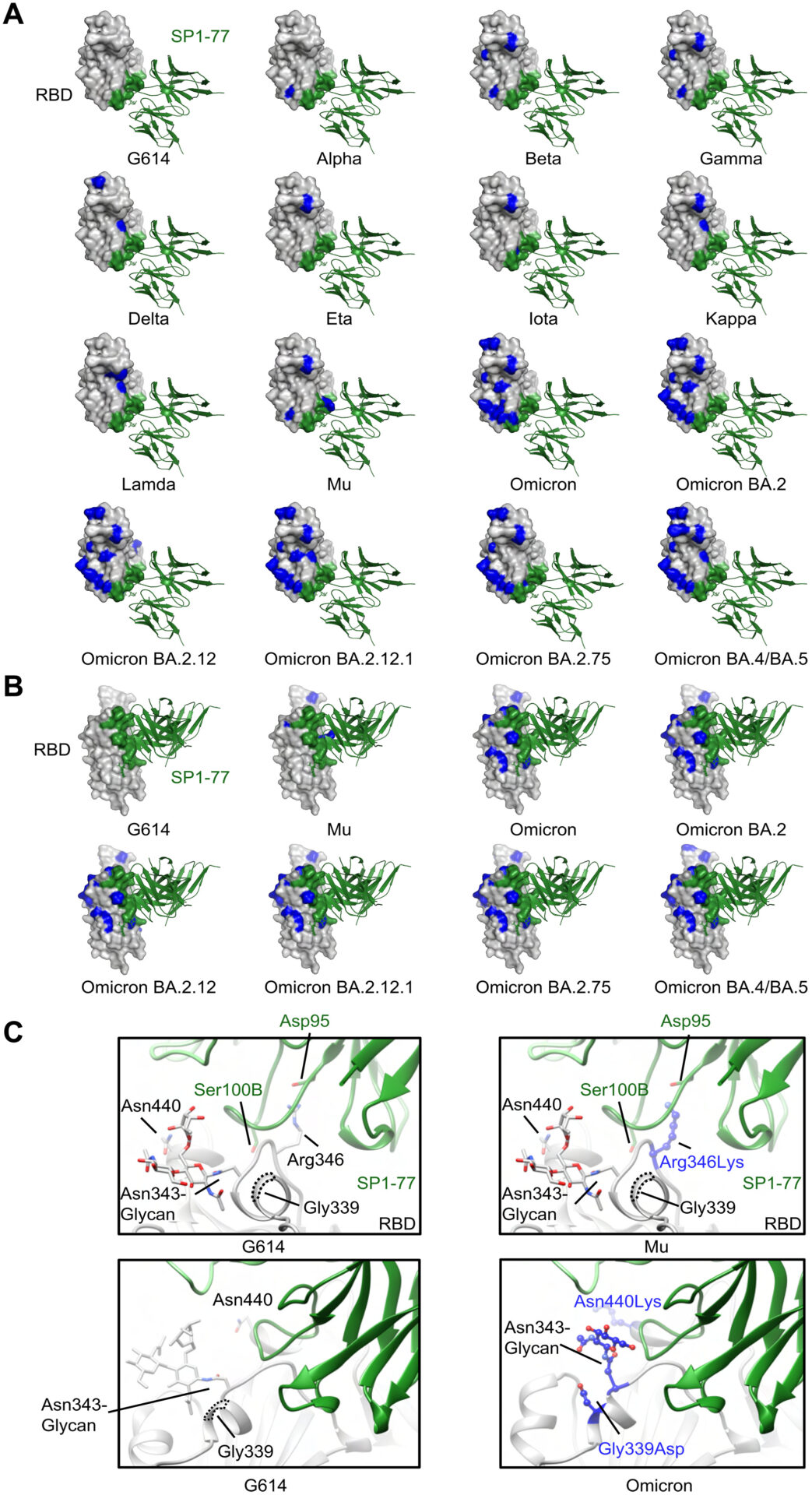
Modeled SP1-77 binding site on various SARS-CoV-2 variants.
(A) The potential footprint of SP1-77 on the modeled RBDs from different SARS-CoV-2 variants in a top view. The RBD was shown in surface representation in gray with the SP1-77 footprint highlighted in green and the mutations in each variant in blue. The Fv region of SP1-77 was shown in ribbon diagram in green. Most mutations on spike variants were not located at the SP1-77 footprint. (B) Side view of a selected panel from A. (C) Structural comparison of the SP1-77 binding interface among the RBDs of wildtype G614, Mu and Omicron variants. The conservative mutation Arg346Lys in Mu preserved the salt bridge between the residue 346 in the RBD and the SP1-77 Asp95. The mutations in Omicron variant reconfigured the local conformation near the N343-glycan, which is on the edge of SP1-77 footprint. The RBD was colored in gray, the SP1-77 heavy chain was colored in green and the light chain was colored in light green. Mutations in the Mu and Omicron variants were shown in stick and ball model in blue.
As the COVID-19 pandemic wears on, newer variants of the SARS-CoV-2 coronavirus have been evolving ways to evade the antibodies we make in response to vaccines or prior infections. As a result, we’ve seen breakthrough cases, antibody treatments that once worked have also become less effective over time. Scientists have been searching for an antibody that would be broadly neutralizing — able to fight off any virus variant that might emerge.
An antibody developed by researchers at Boston Children’s Hospital now seems to fit the bill. In lab tests, it neutralized all currently known SARS-CoV-2 variants of concern, including all Omicron variants.
“We hope this antibody will prove to be as effective in patients as it has been in pre-clinical evaluations thus far,” says Dr. Frederick Alt of the Program in Cellular and Molecular Medicine at Boston Children’s, a senior investigator on the study. “If it does, it might provide a new therapeutic and also contribute to new vaccine strategies.”
Creating and testing multiple antibodies
Led by Dr. Alt and Dr. Sai Luo at Boston Children’s, the team first modified a mouse model the Alt lab created to search for broadly neutralizing antibodies to HIV, another virus that frequently mutates. These mice essentially have built-in human immune systems. The modified model mimics — and refines — the trial-and-error process our own immune systems use to create increasingly effective antibodies when we encounter an invader.
The researchers first inserted two human gene segments into the mice. This pushed their immune cells to quickly produce a diverse repertoire of antibodies resembling those we ourselves might make. The team then exposed the mice to the SARS-CoV-2 spike protein, the main protein targeted by our antibodies and current vaccines, from the original Wuhan-Hu-1 coronavirus strain.
In response, the mice produced nine different “families” of antibodies that bound to the spike. With collaborators at Duke University, Drs. Alt and Luo then tested the effectiveness of these antibodies.
Antibodies in three of the nine families strongly neutralized the original Wuhan-Hu-1 virus. But one antibody family, and especially an antibody dubbed SP1-77, showed much broader activity, neutralizing Alpha, Beta, Gamma, Delta, and all previous and current Omicron strains.
A new way to neutralize the virus
What made SP1-77 so good at neutralizing the virus? Structural biology studies by collaborating teams led by Drs. Bing Chen and Jun Zhang at Boston Children’s and Dr. Barton Haynes at Duke showed that this antibody works in a unique way.
In order to infect us, SARS-CoV-2 must first attach to ACE2 receptors on our cells. Many of the antibodies we make in response to vaccines, as well as antibodies used to treat COVID-19, block this binding. They do so by attaching to the spike’s receptor-binding domain (RBD) at specific locations. The SP1-77 antibody also binds to the RBD, but in a different way that does not prevent the virus from binding to ACE2 receptors.
So how would it protect us? Well, once the virus has bound to ACE2, it must complete a final step: fusing its outer membrane with the membrane of our cells. This throws the door open to infection. Using a novel live-cell imaging platform, Drs. Alex Kreutzberger and Tomas Kirchhausen of Boston Children’s showed that SP1-77 blocks this step.
“SP1-77 binds the spike protein at a site that so far has not been mutated in any variant, and it neutralizes these variants by a novel mechanism,” notes Dr. Kirchhausen. “These properties may contribute to its broad and potent activity.”
Better COVID-19 vaccines and treatments?
The researchers have applied for patents on both the antibodies and the mouse model used to produce them. They hope to see their work developed commercially.
“It was a pleasure to work with this dedicated collaborative team whose diverse expertise allowed SP1-77 to be brought to light,” says Alt. “We look forward to using our models to discover antibodies with therapeutic potential against other newly emerging pathogens.”
Original Article: Powerful new antibody neutralizes all known coronavirus variants
More from: Boston Children’s Hospital | Duke University
The Latest Updates from Bing News
Go deeper with Bing News on:
Coronavirus neutralizing antibody
- Feed has no items.
Go deeper with Bing News on:
SP1-77
- Feed has no items.